论文地址:Evaluating the representational power of pre-trained DNA language models for regulatory genomics
gLM-eval:评估glm在各种zero-shot下游任务表现
Abstract
genomic language models (gLMs)的涌现提供了一个无监督的方法来学习非编码基因组中的顺式调控模式。先前的评估已经展现了预训练的gLMs可以提高调节基因组任务的预测能力,尽管这样的微调效果很好,但确定gLM表征是否体现了对顺式调节生物学的基本理解仍然是一个悬而未决的问题。这里我们用预训练模型来解释跨越DNA和RNA调控的细胞类型特异性功能基因组学数据。研究结果表明,与使用单热编码序列的传统机器学习方法相比,目前的glm并没有提供实质性的优势。这项工作强调了当前glm的一个主要局限性,提出了非编码基因组的传统预训练策略的潜在问题
Introduction
预训练蛋白质模型的表征用途很多,可以预测蛋白质结构、非同义突变效应,设计新的蛋白质序列,研究蛋白质进化关系
而对基因组DNA序列进行预训练的模型加速了对非编码基因组中的功能元素理解,gLMs原则上可以帮助理解转录因子(TFs)控制顺式调节元件(CREs)活性的复杂协调
输入是DNA序列,分词方法有单核苷酸和k-mer两种,基础架构有transformer,多头注意力或者一些变体,或是残差连接的卷积网络;数据比较丰富,比如有单个物种的基因组,多个物种的基因组或是基因组的特定区域,比如未翻译区域(UTRs)、pre-mRNA、启动子、编码区、非编码RNA或是保守位点
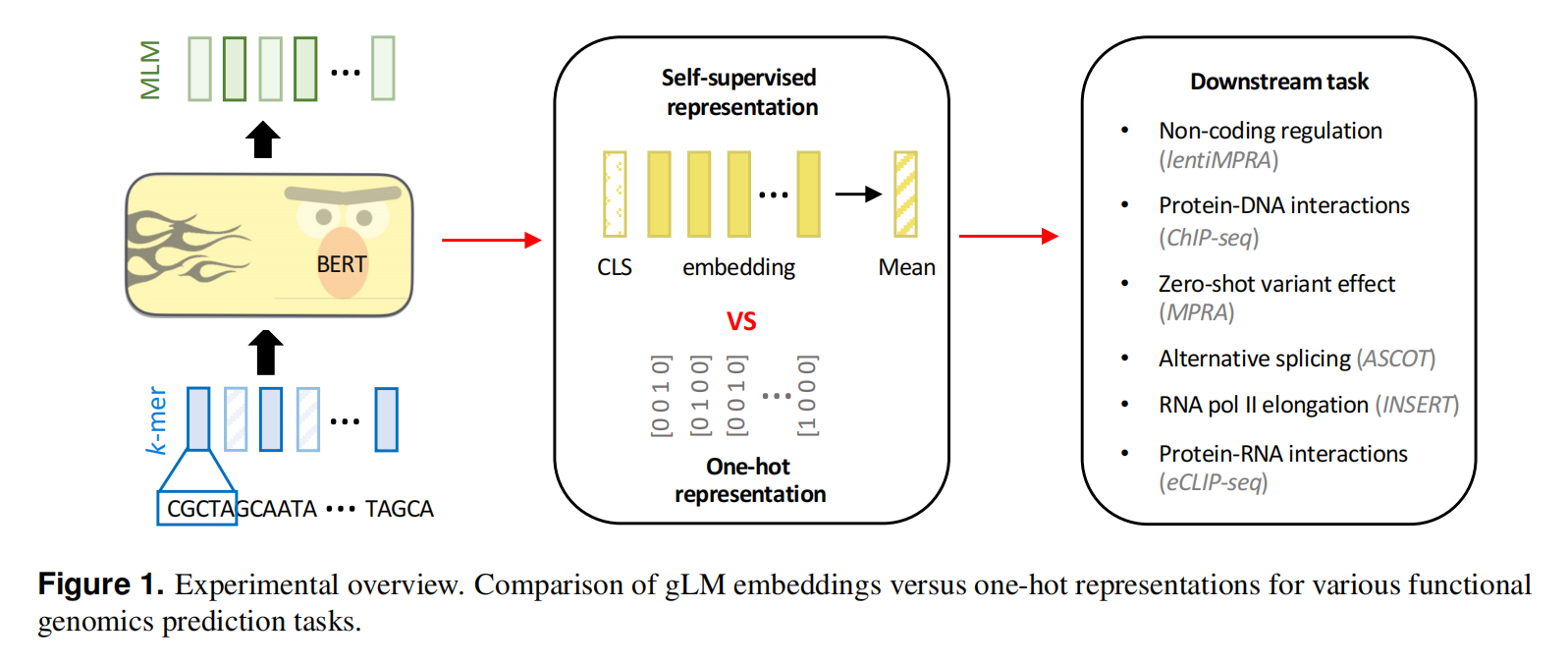
这里作者评估了六种主要的功能基因组预测任务,涵盖了在DNA和RNA水平上的顺式调控复杂性,结果显示目前的预训练gLMs并没有比传统的one-hot表示+深度学习强多少。同时,对功能的基因组数据进行监督学习的foundation models可以更好的迁移到其他功能基因组数据上。还有结果显示,当前的gLM的预训练方案难以理解细胞类型特定功能元素,还不能实现真正的基础模型的效果
Results
Task 1: Predicting cell-type specific regulatory activity from lentiMPRA data
通过考虑两种细胞类型,即HepG2和K562,我们可以评估预训练的gLM表示是否捕获了细胞类型特异性的CRE活性,输入DNA序列,预测一个CRE的标量
使用CLS token或是mean embedding,使用CNN作为Baseline,用倒数第二层的full embedding和one-hot序列做比较,还是用了一个有监督的foundation model Sei,使用one-hot sequence的ResNet model,以做到全面而公平的比较
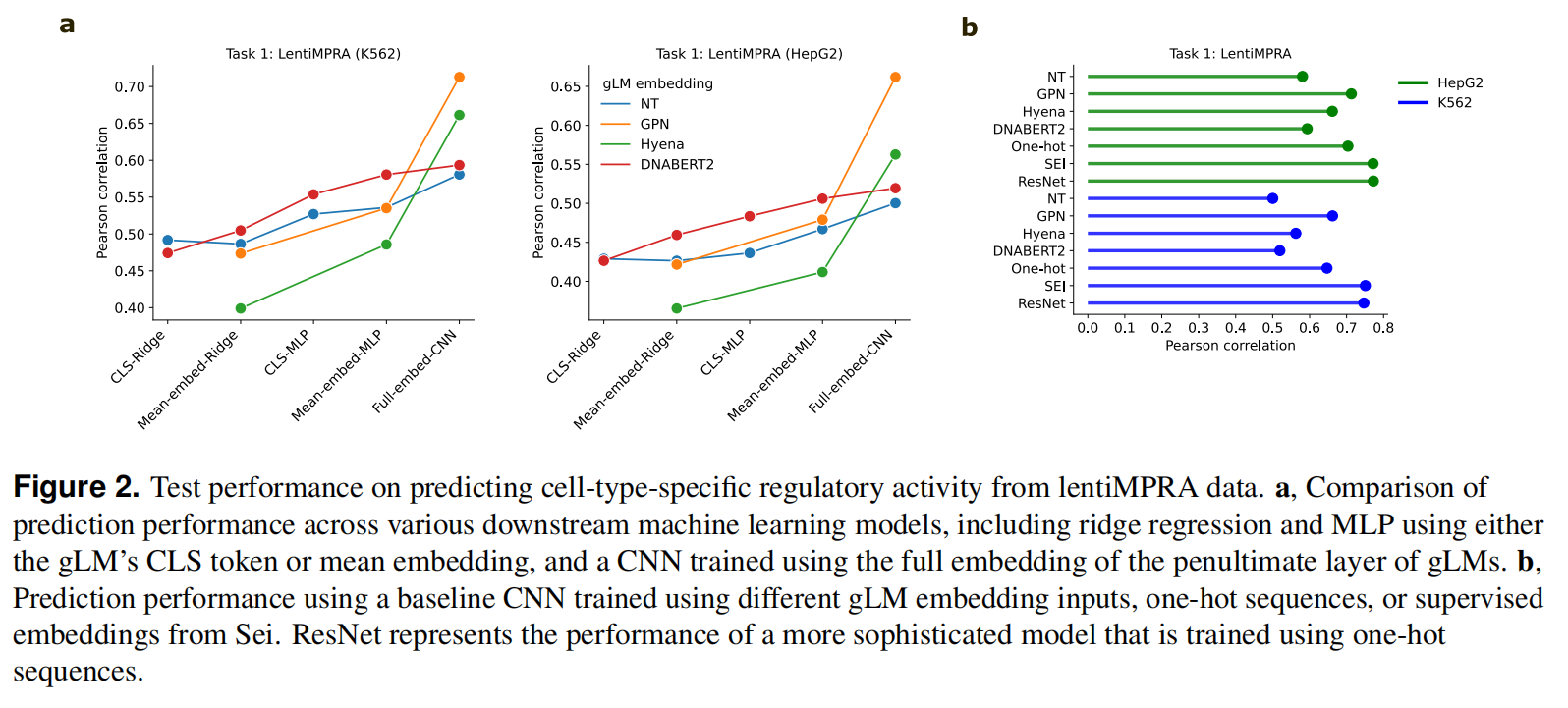
fig2 b可以看到很多情况下,embedding方法甚至不如one-hot
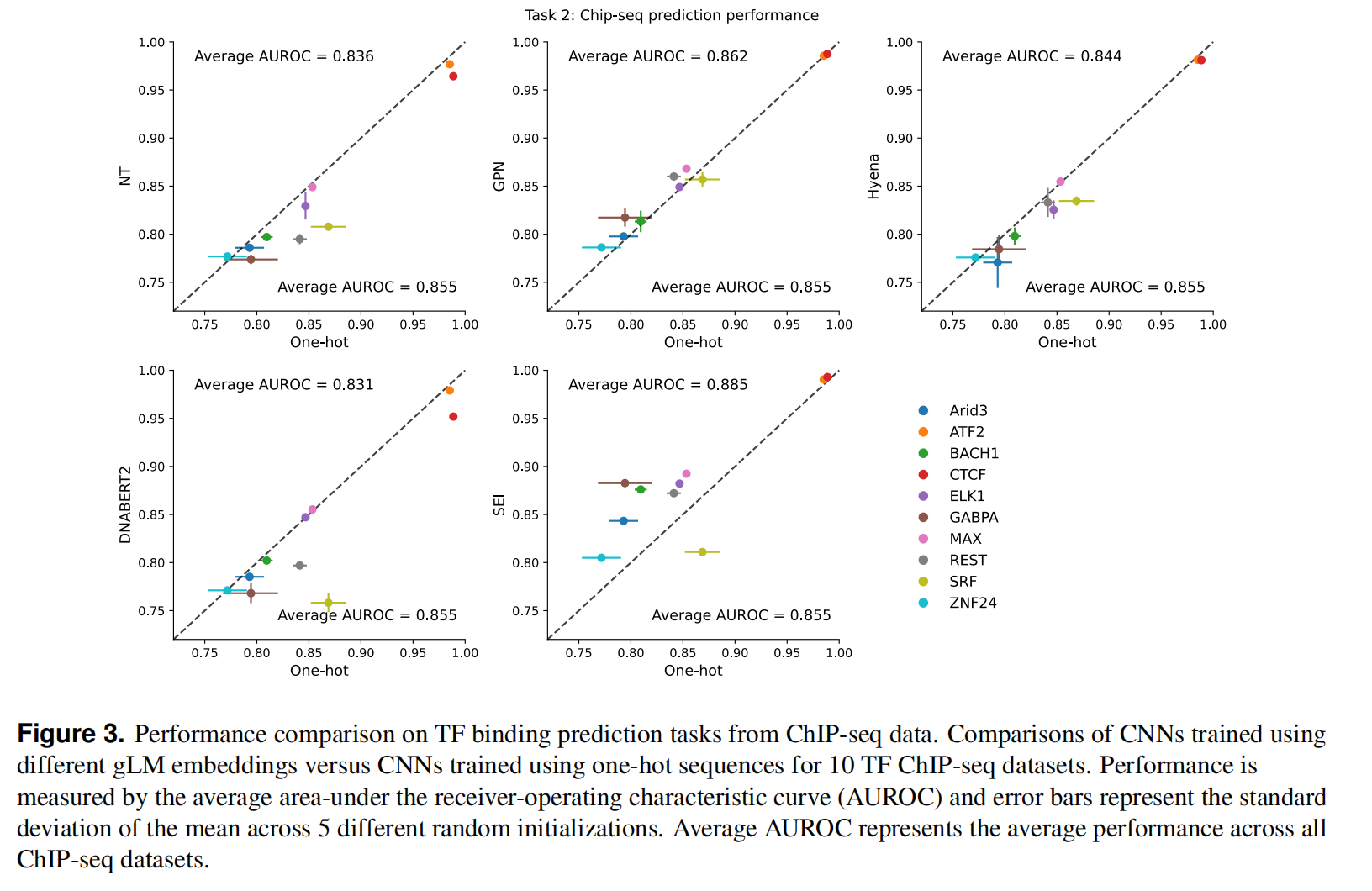
Task 2: Predicting TF binding sites from ChIP-seq data
TF binding是一个细胞类型特异性的现象,而标准的语言建模方法是没有做到cell-type感知的,一个可能的原因是lentiMPRA预测任务在训练时丢失了关键motif
Task 3: Zero-shot variant effect prediction with MPRA data
预测非编码突变
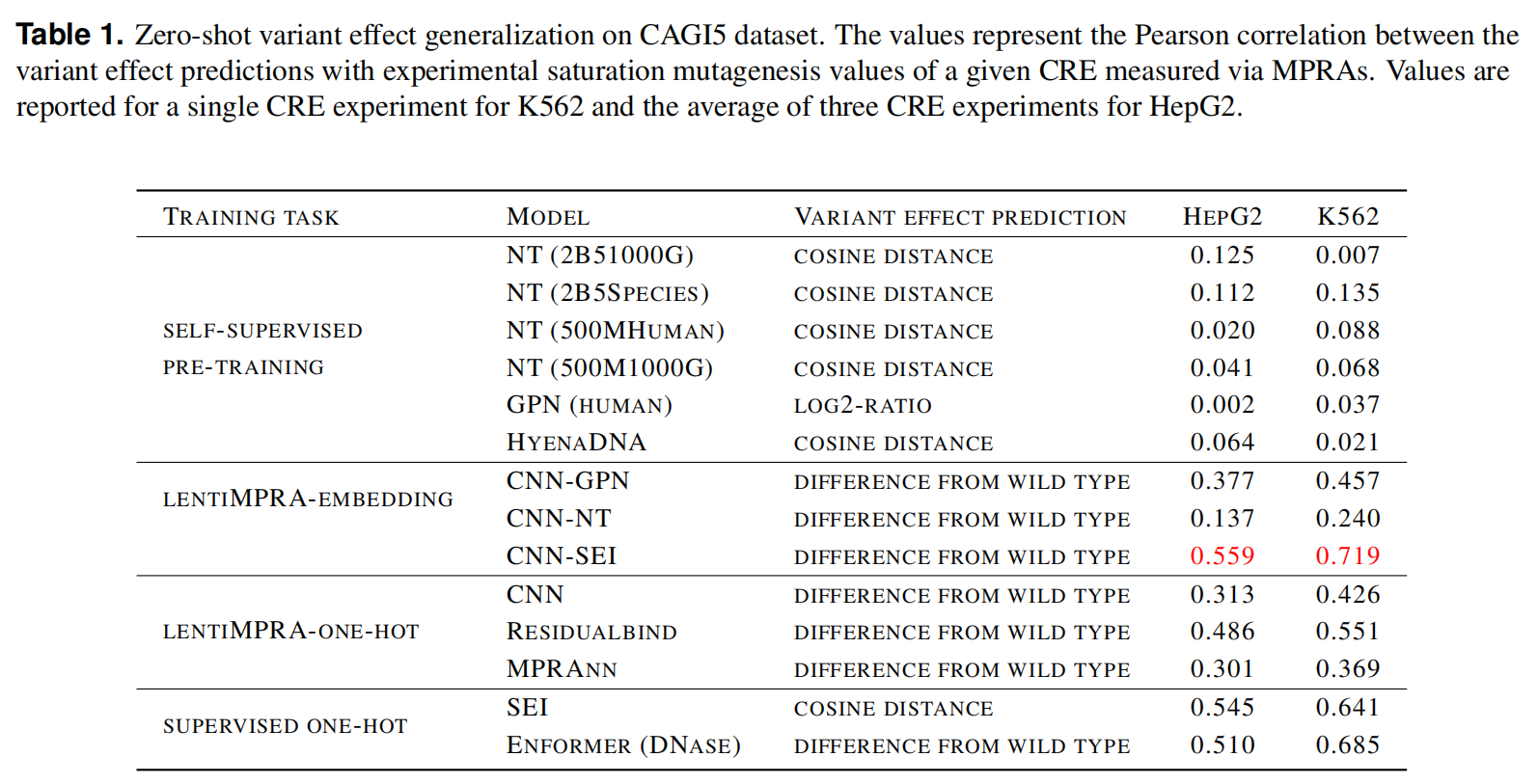
使用gLM嵌入在lentiMPRA数据上训练的cnn相对于预先训练的同行产生了明显更好的性能
Task 4: Predicting alternative splicing from RNA-seq data
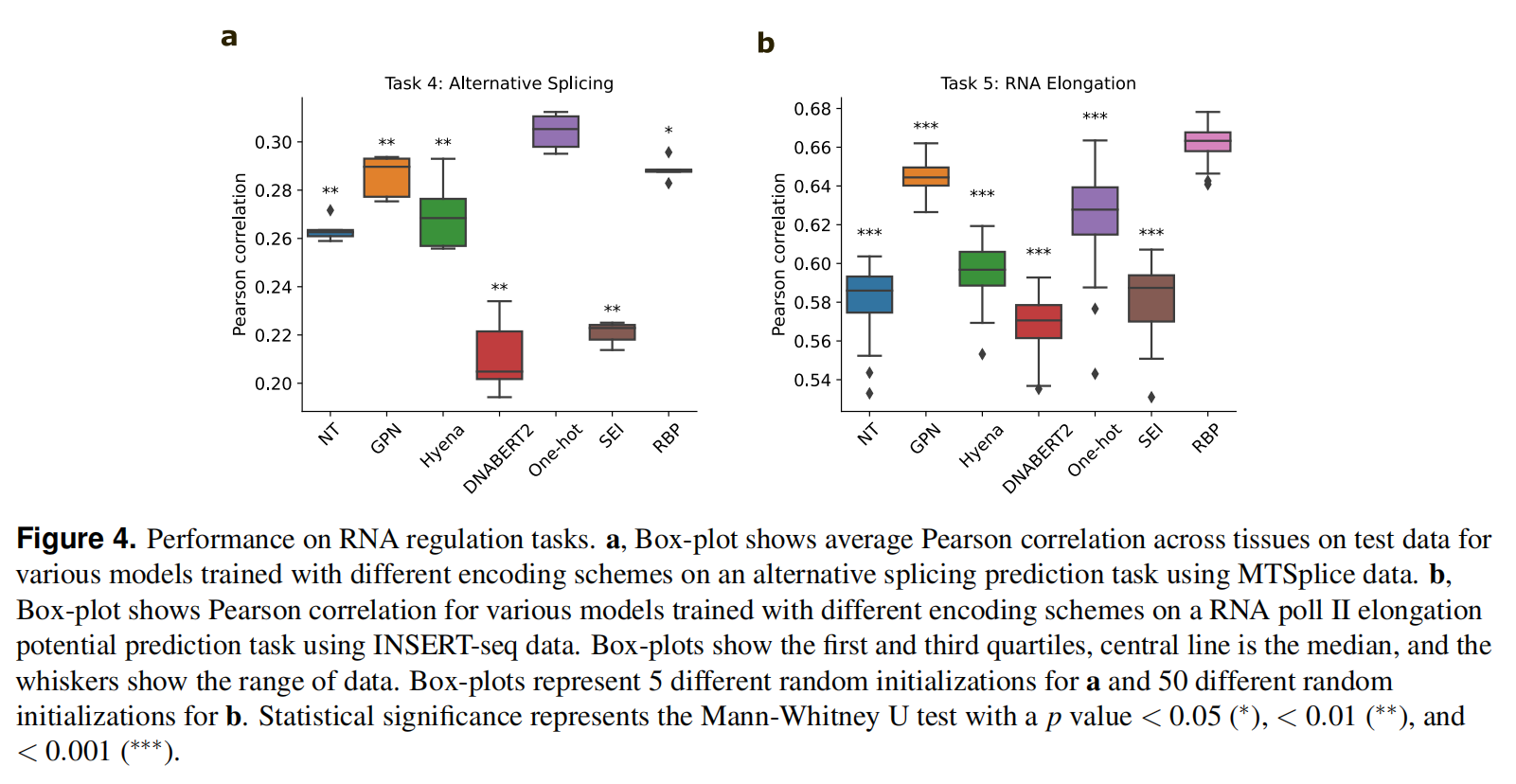
先前的研究表明,Nucleotide-Transformer和GPN已经学习了与基因定义和剪接位点相关的特性,使用ASCOT数据集测试
Task 5: Predicting RNA pol II elongation potential from INSERT-seq data
以nt RNA序列作为输入,并预测RNA pol II的延伸,使用INSERT-seq dataset,从fig4可以看到预训练模型并不一定能取得更好的效果
Task 6: Predicting RNA-binding protein binding with eCLIP-seq data
RBP对于不同的RNA加工阶段都是必不可少的,使用eCLIP-seq(增强染色质免疫沉淀测序)数据集检测了gLMs预测RBP结合位点的能力
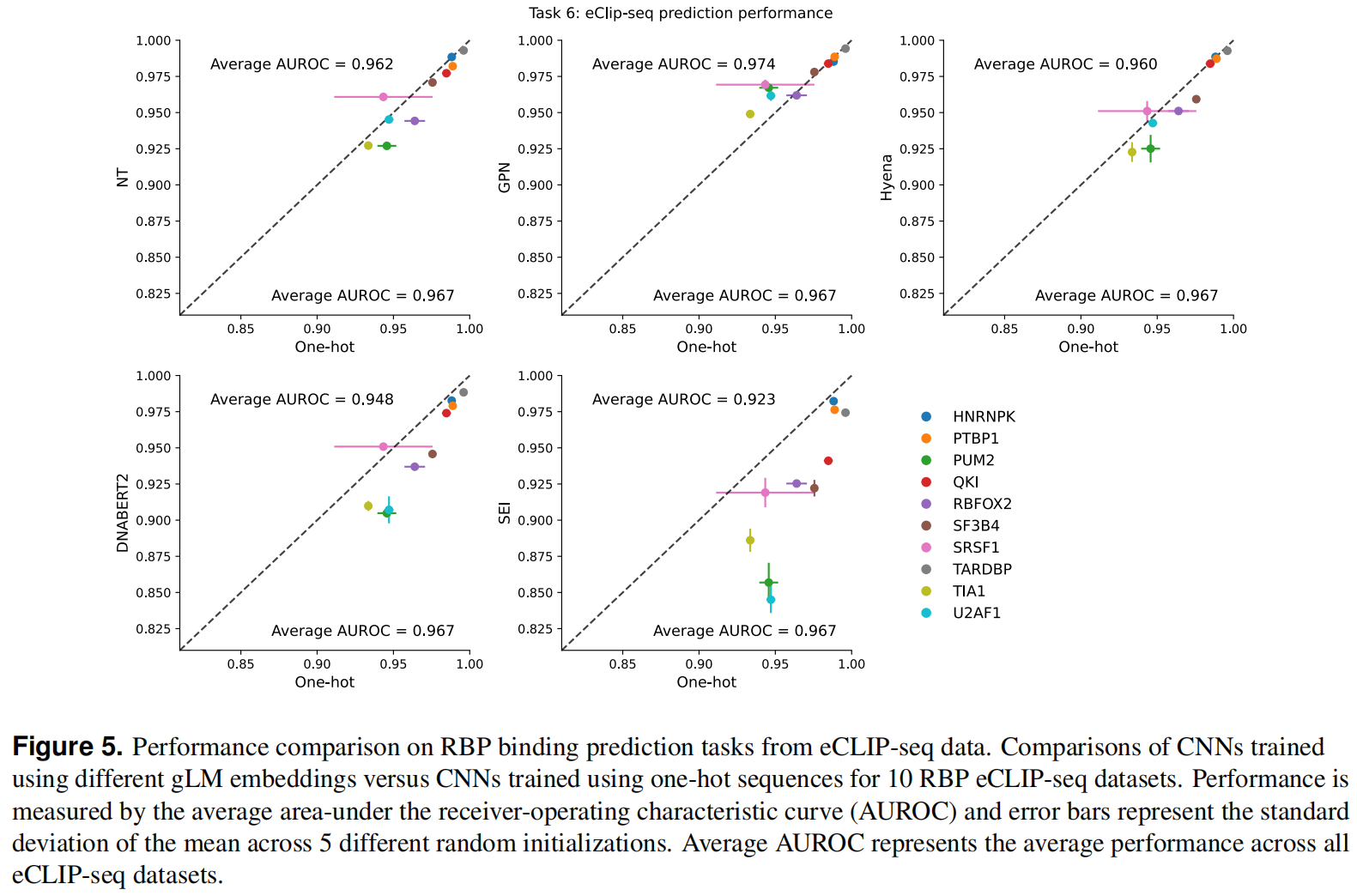
Uncovering cell-type-specific motifs learned by gLMs is challenging
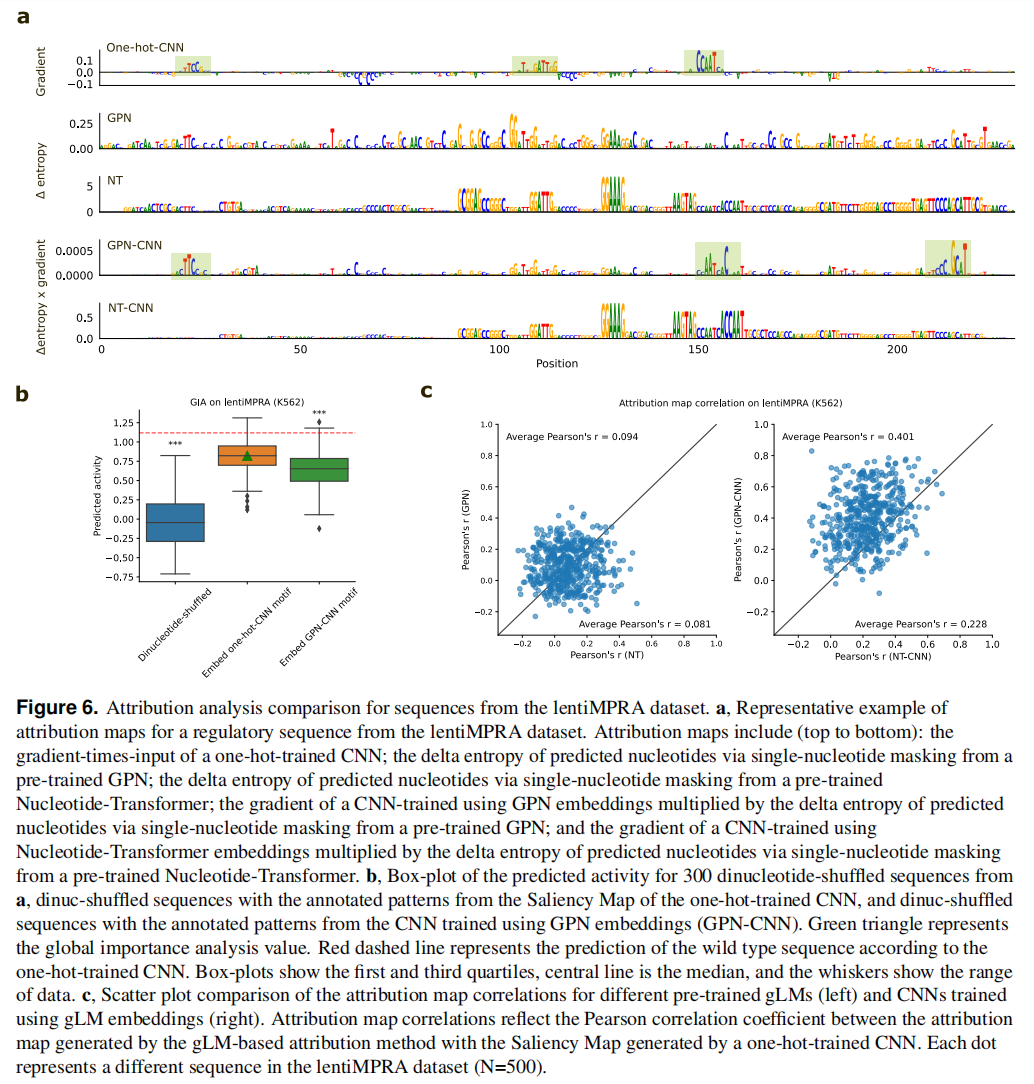
Discussion
与标准的one-hot序列相比,gLM表示几乎没有提供任何优势
选择不对每个下游任务上的gLM的权重进行微调,虽然gLM的性能可能会通过微调而提高,但本研究的范围是严格衡量在训练前所学到的顺式调节生物学知识
将当前的预训练的任务MLM扩展到整个基因组,难以捕获非编码基因组中有意义的表征,目前尚不清楚使用标准语言建模目标(即MLM或CLM)预训练的glm的持续缩放是否会最终会涌现s


